Software for working with data obtained using the GeneValue™ NSCLC Solution. Does not require user bioinformatics training.
GeneValue™ Somatic Software
Access to the GeneValue™ Somatic Software is free with the purchase of GeneValue™ NSCLC Solution.

GeneValue™ Somatic Software provides high-throughput sequencing analysis, quality control and annotation of genetic variants. Since it is deployed in the cloud, the software does not require the user's own computing power.
Simple workflow
Importing data and metainformation
Data analysis and quality control
Annotation and visualization
Report generation
Pipeline DualStream

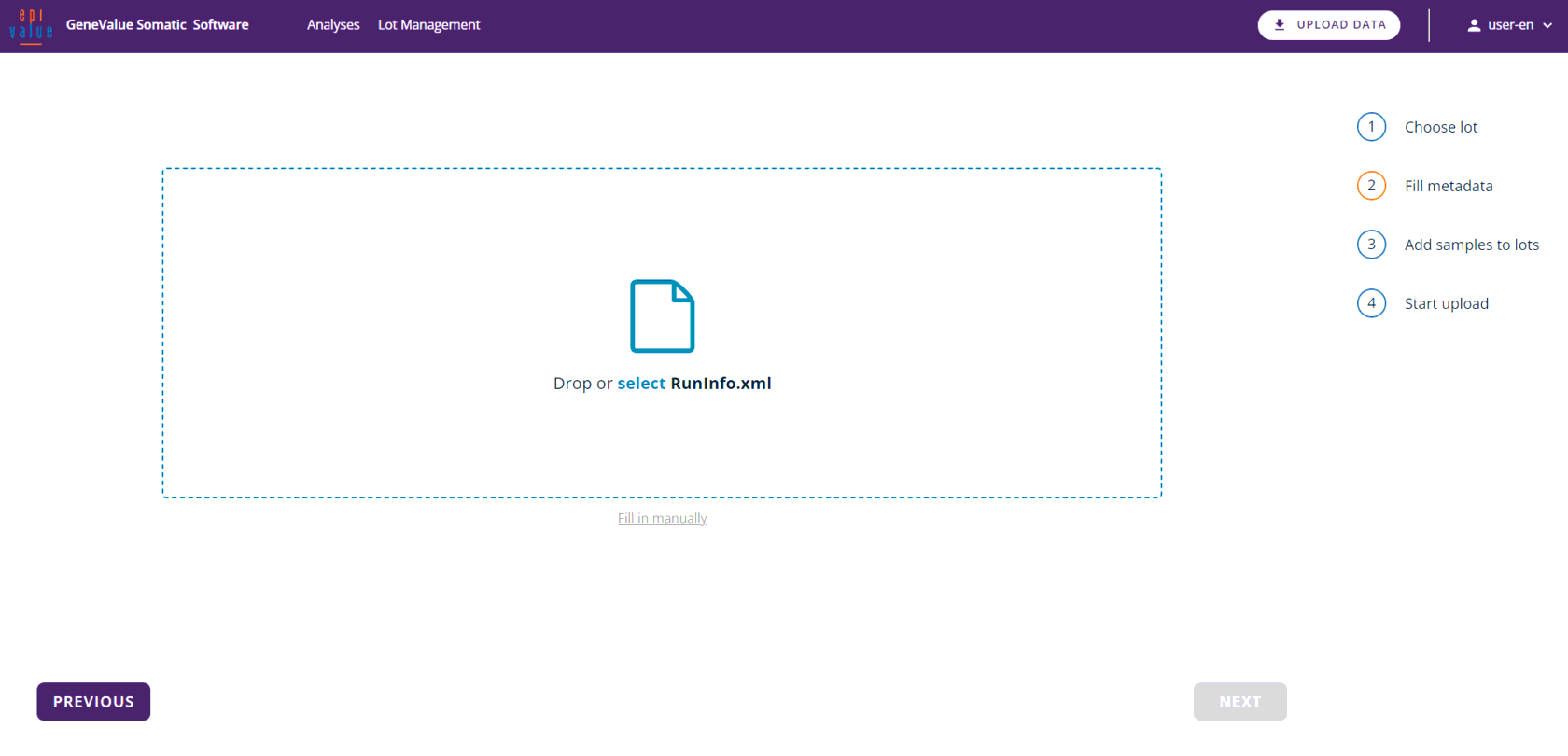
The data upload interface allows the importation of sequencing results and initialisation information. The assay can be run simply by entering the reagent batch number. All files can be uploaded via the "select" button or simply by dragging and dropping the relevant file into the active field.
Importing data and metainformation
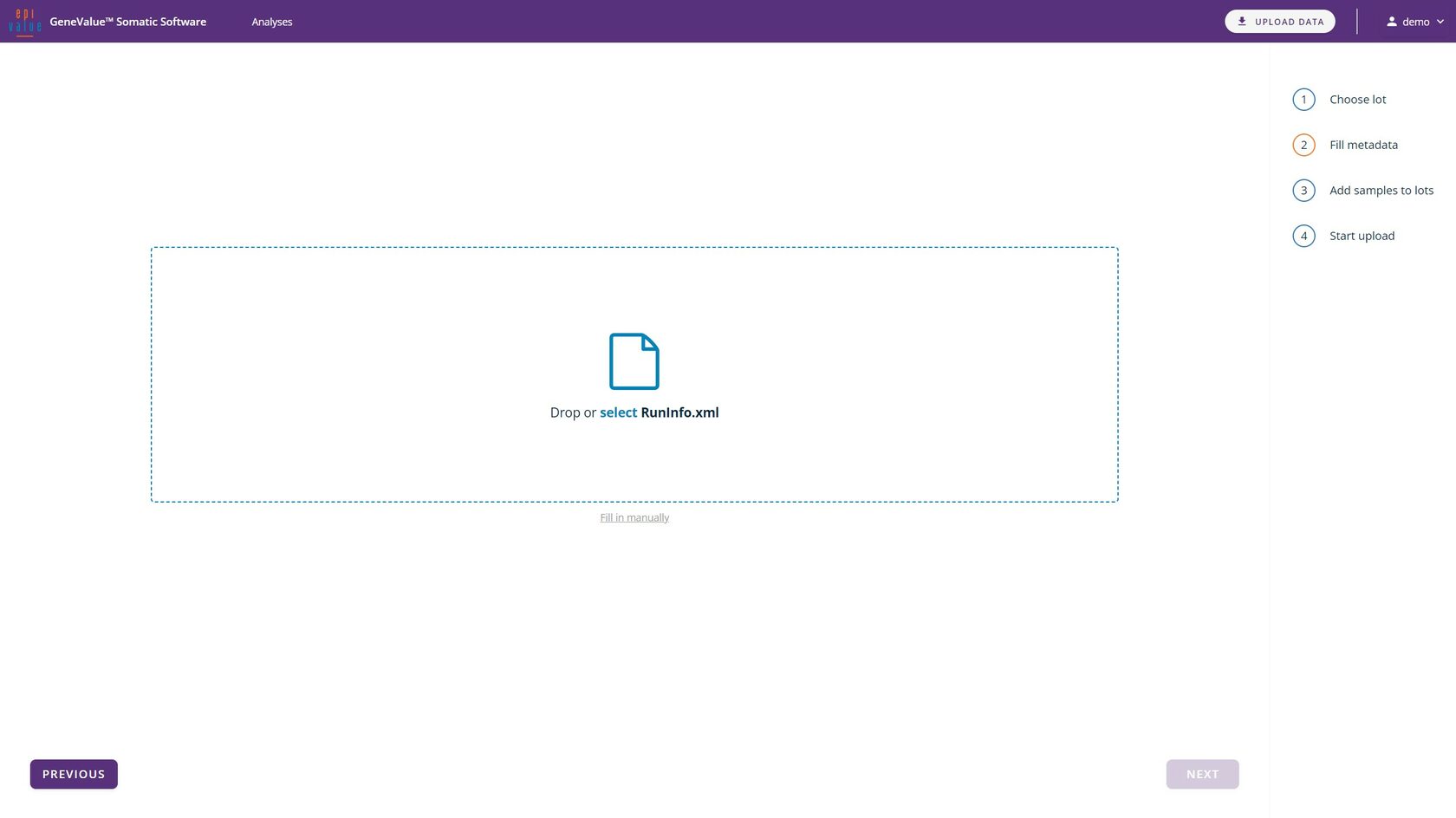
The data upload interface allows the importation of sequencing results and initialisation information. The assay can be run simply by entering the reagent batch number. All files can be uploaded via the "select" button or simply by dragging and dropping the relevant file into the active field.
Importing data and metainformation
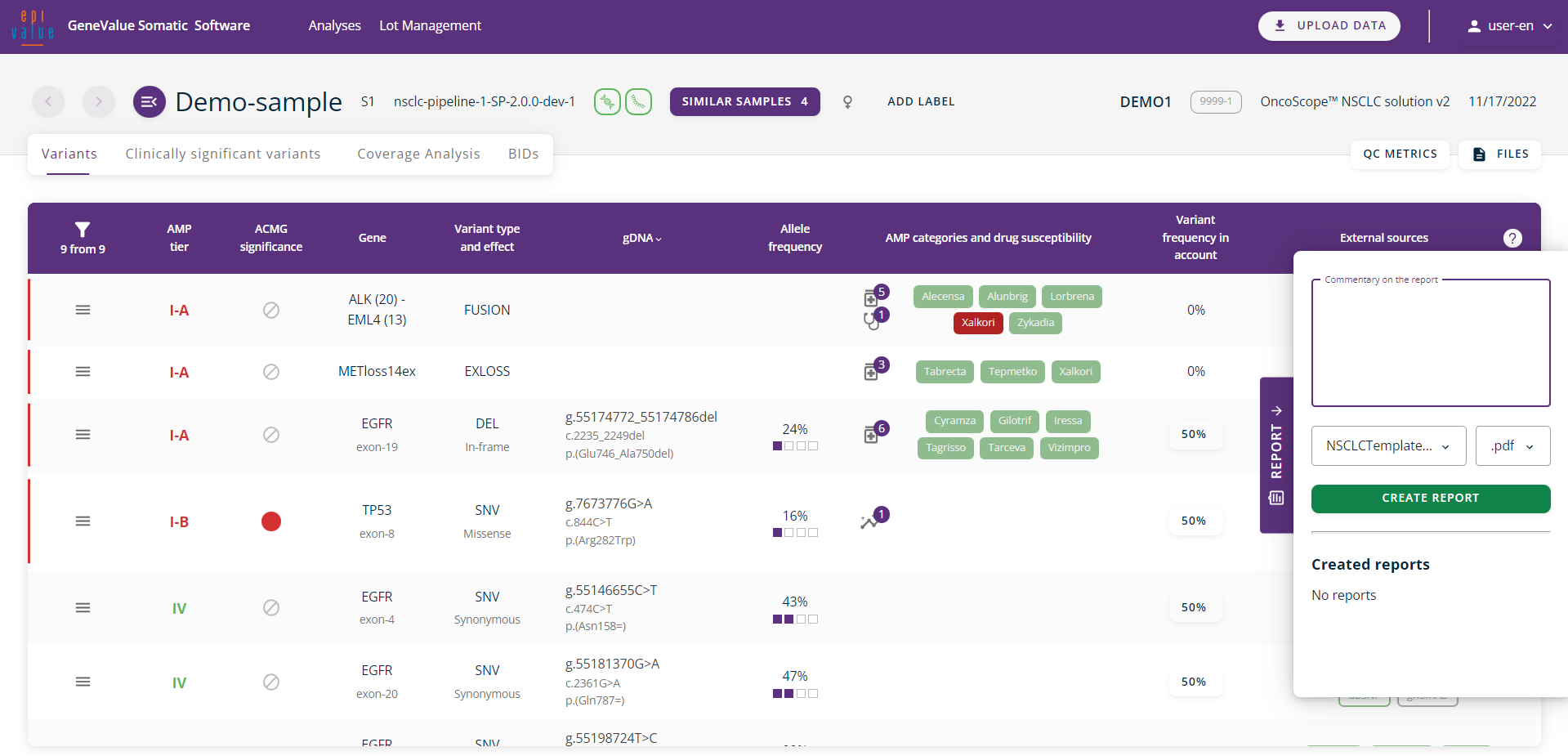
All found variants are visualised on the main working tab, indicating AMP tier, ACMG significance, the gene, variant type and effect, the HGVS nomenclature, allele frequency, AMP categories and drug susceptibility, variant frequency by account and links to external sources.
Data analysis
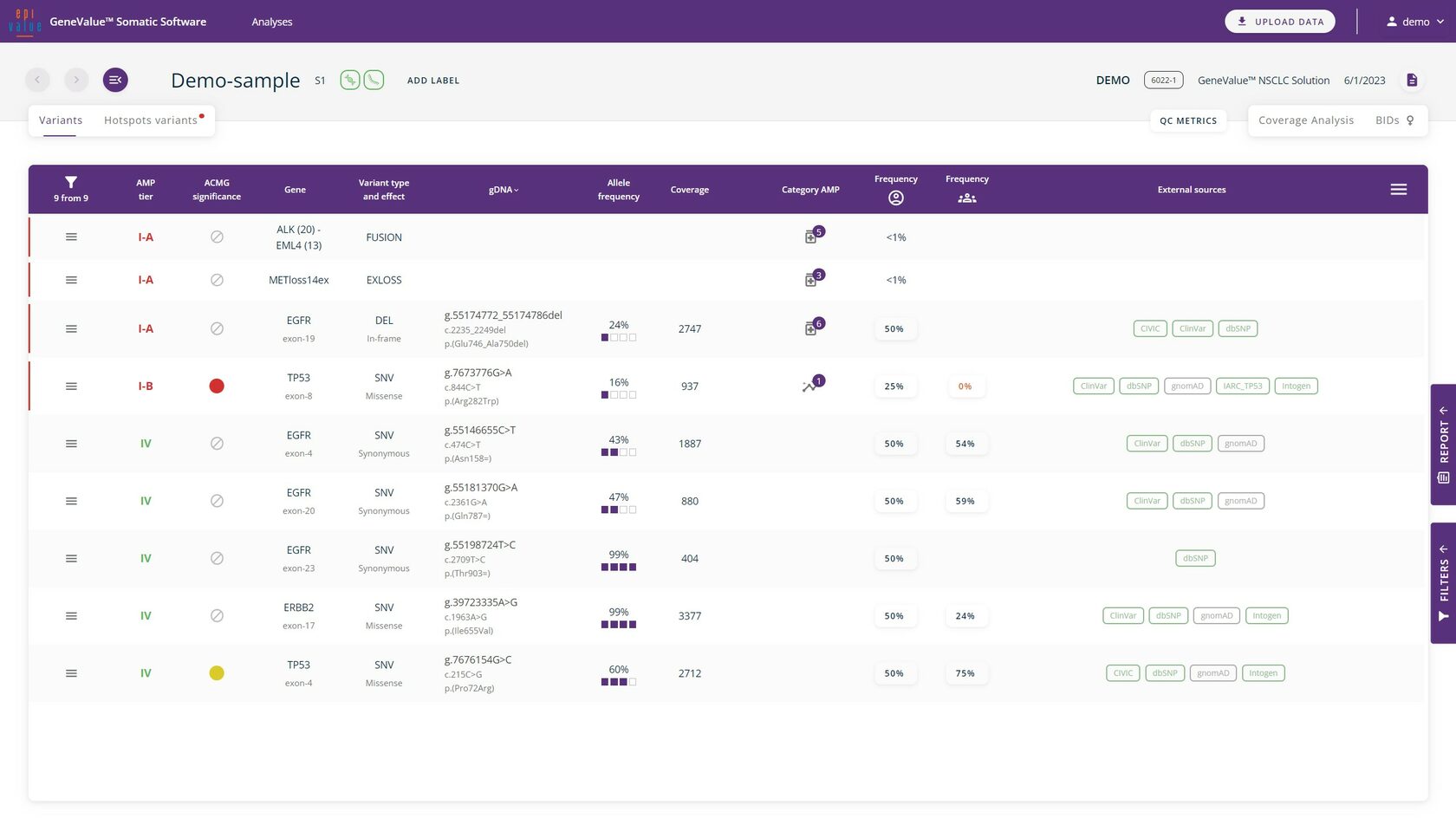
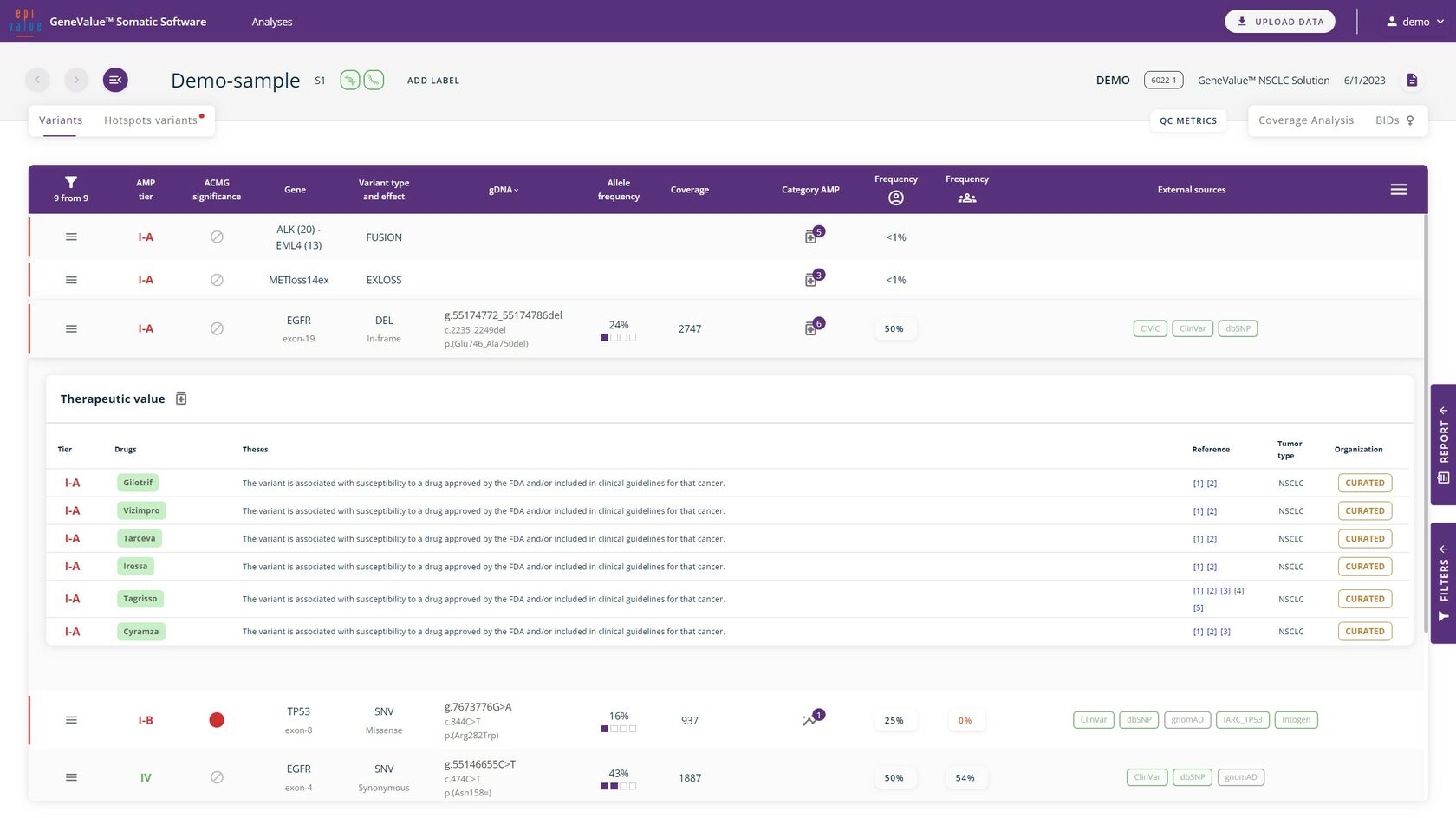
The most commonly occurring variants are annotated according to ACMG or AMP requirements. For rare, non-annotated variants, it is possible to request an annotation from our specialists.
Annotations
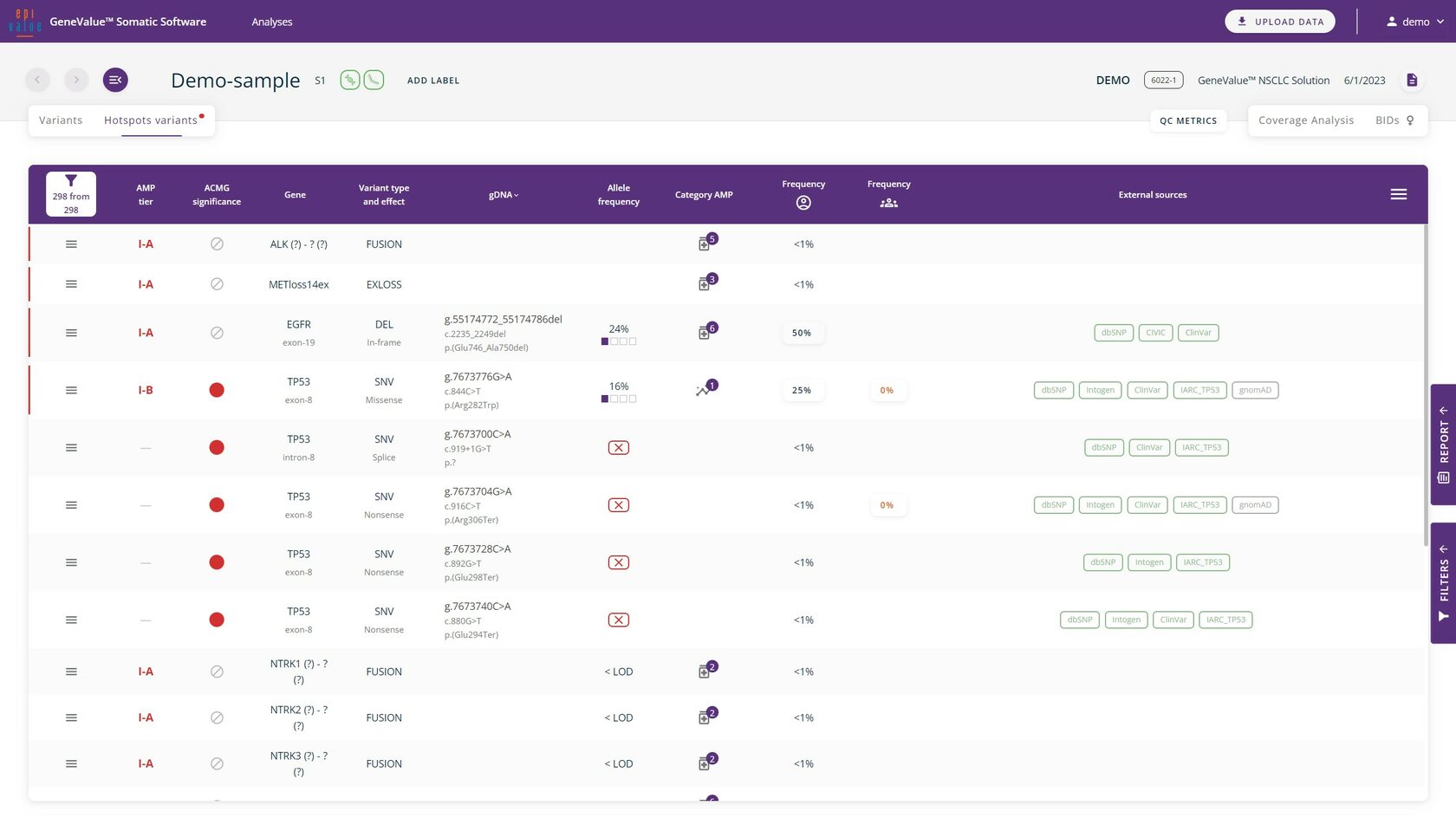
All hotspots defined for the used panel are rendered in a separate "Assay hotspots" tab. The double control performed in this tab allows elimination of variants in hotspots.
Clinically significant variants (hotspots)
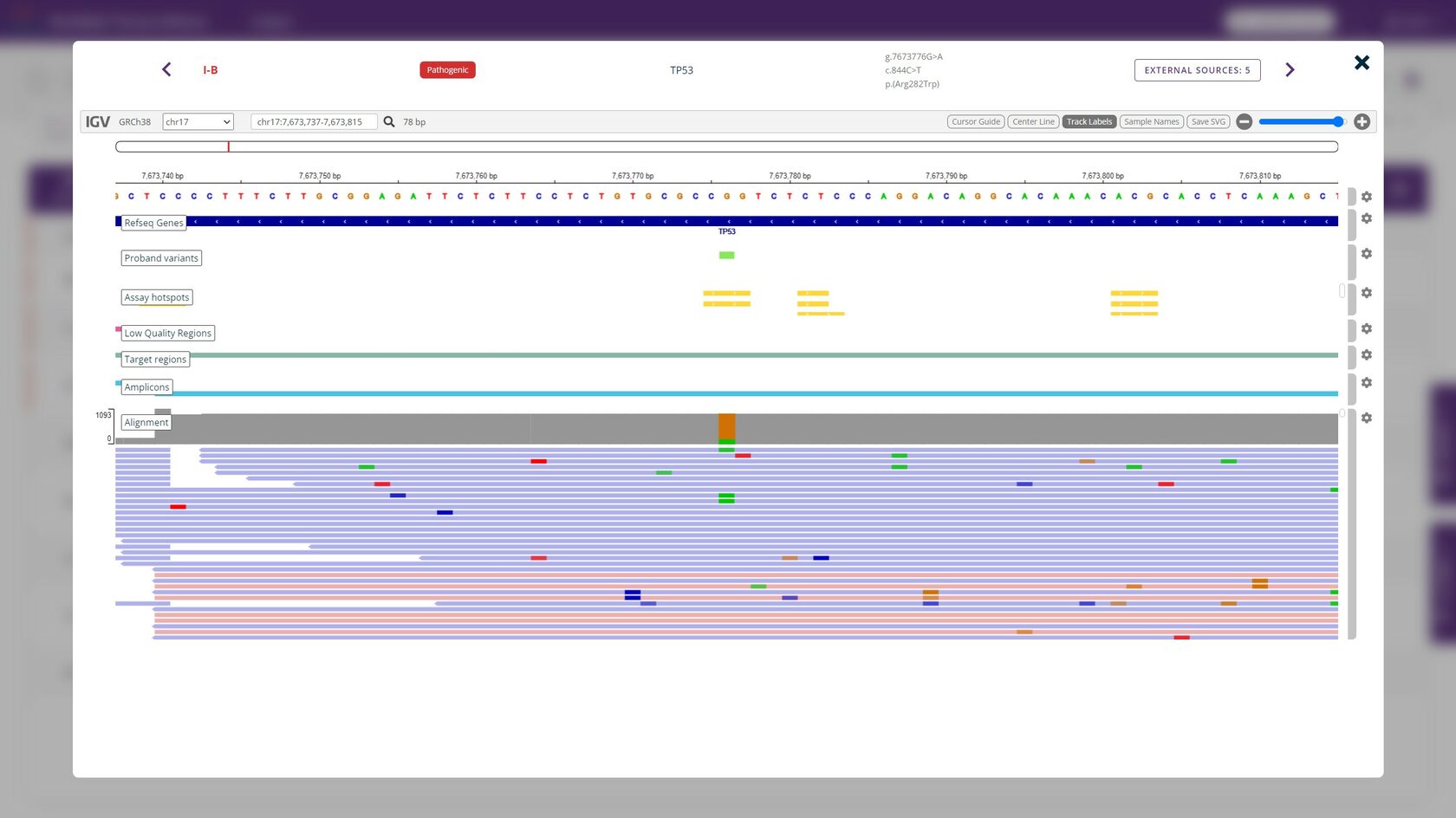
The quick visual evaluation of results by the built-in IGV browser is useful when monitoring hotspot variants in regions with low coverage.
IGV browser
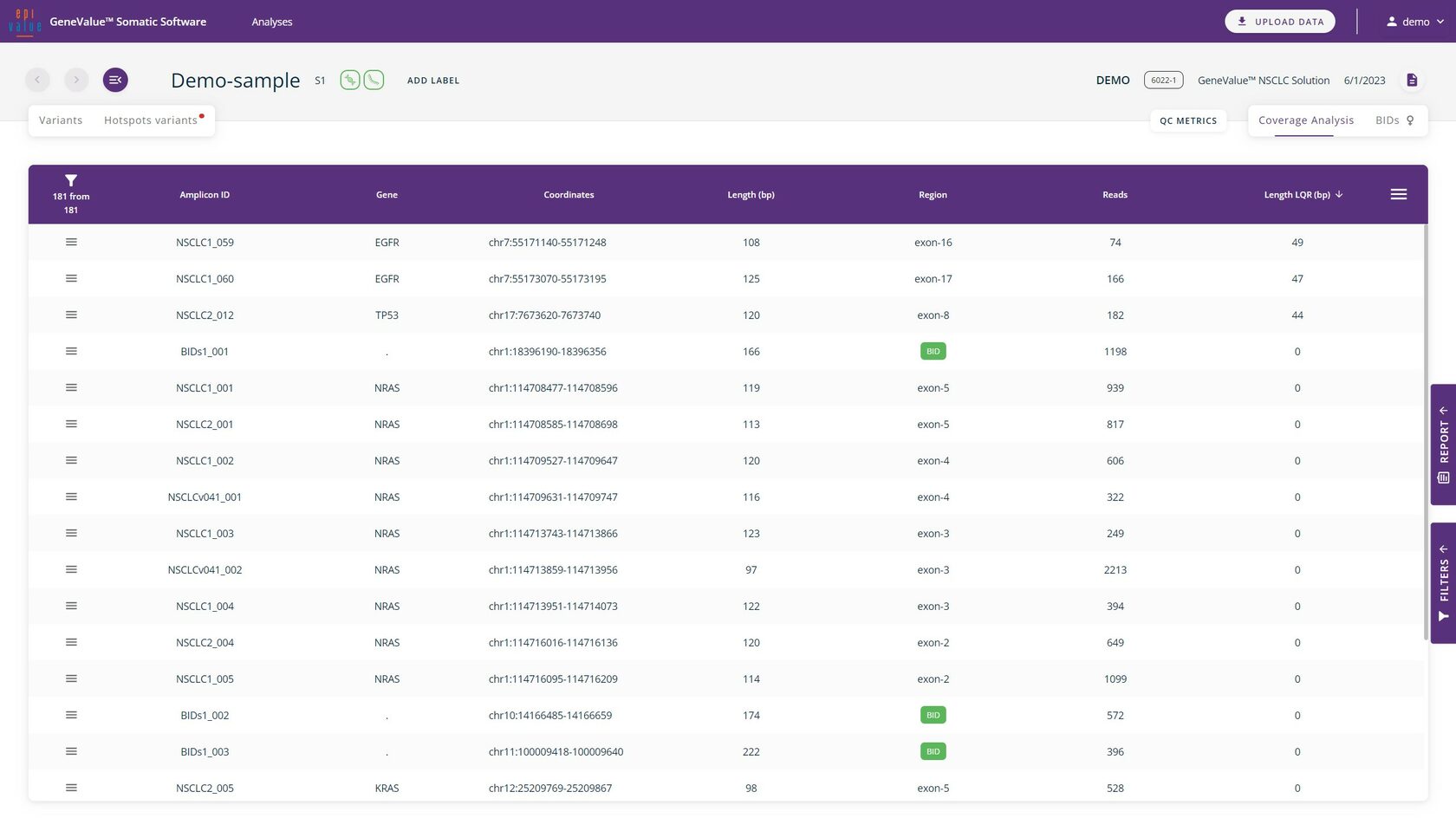
The Coverage Analysis tab displays the number of reads, common length and length of LQR (low quality region) for each amplicon.
The Coverage Analysis
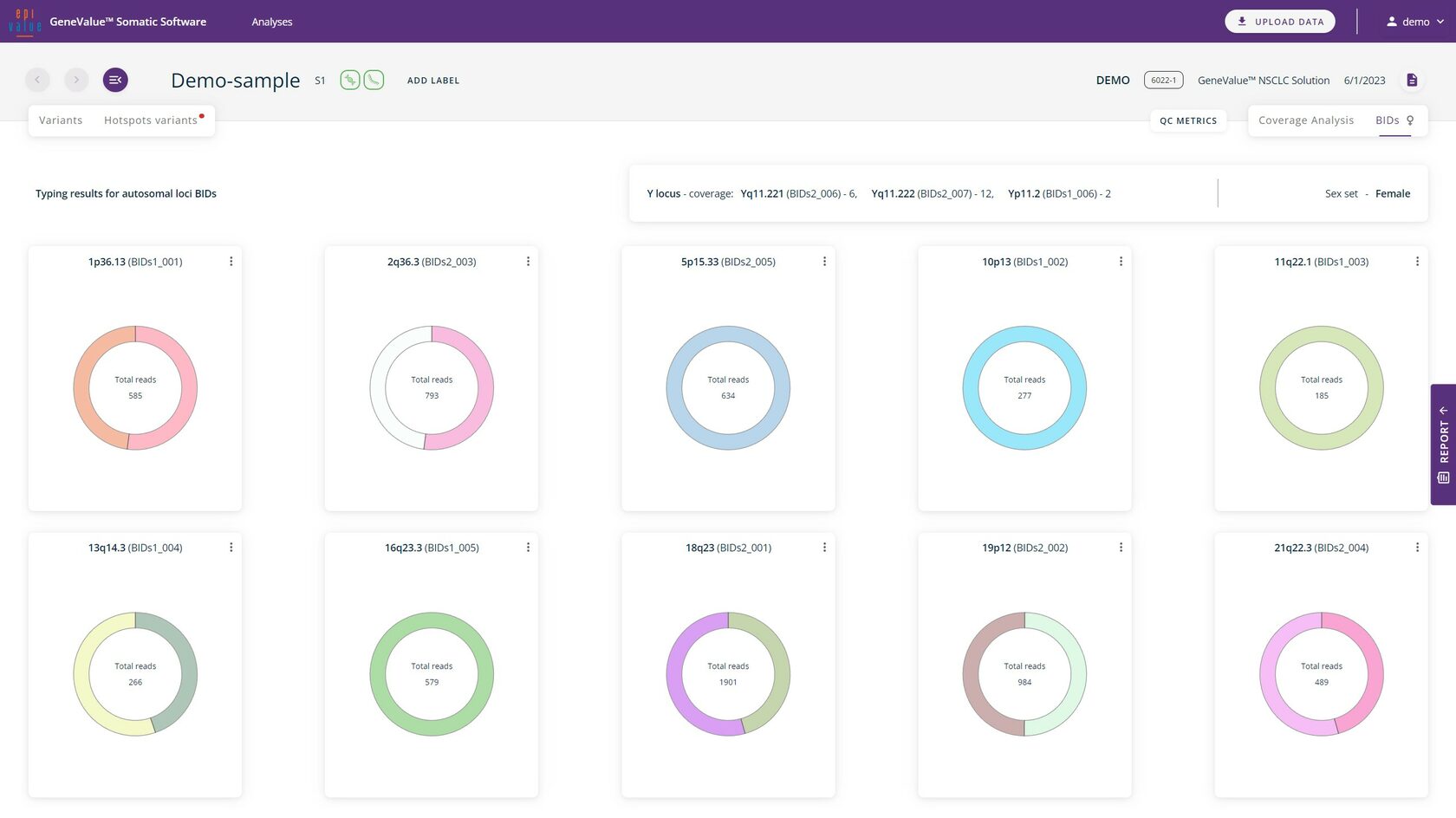
The biological identifiers (BIDs) included in the GeneValue™ NSCLC Solution comprise amplicons on variable regions of the genome. These can be used to detect the presence of sample contamination, as well as for determining the sex.
BIDs
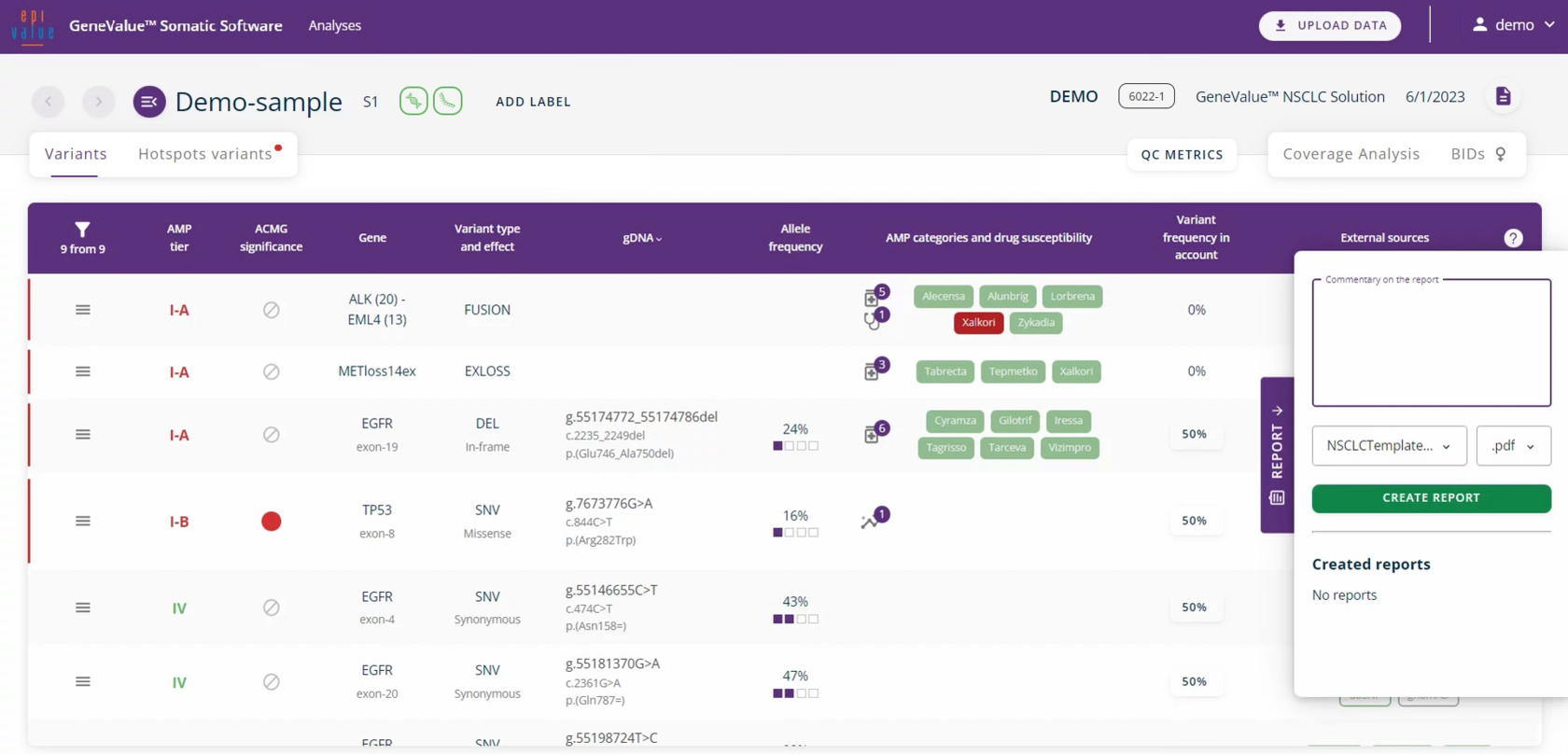
The system generates a report indicating the analysed genes, found variants and quality parameters.
Report
The system generates a report indicating the analysed genes, found variants and quality parameters.
Report