A test system for identifying mutations associated with cystic fibrosis, phenylketonuria and galactosaemia using next-generation high-throughput sequencing.
GeneValue™ Neoscreen Solution
For research use only

Contains library preparation reagents and access to GeneValue™ Germline Software for data analysis.
Product highlights

High sensitivity and specificity
The solution is validated against samples from the National Institute of Standards and Technology (USA); the bioinformatics pipeline is validated in accordance with the recommendations of the Association for Molecular Pathology (USA).
Fully automatic analysis and data quality assessment, including contamination control and sample sex determination in GeneValue™ Neoscreen Solution software.
Does not require bioinformatician
Simple and fast interpretation
Proprietary mutation base annotated according to standards published by the American College of Medical Genetics and Genomics (ACMG). Links to external sources of annotation such as ClinVar, dbSNP and gnomAD databases. Ability to request a qualified annotation of a discovered new variant.


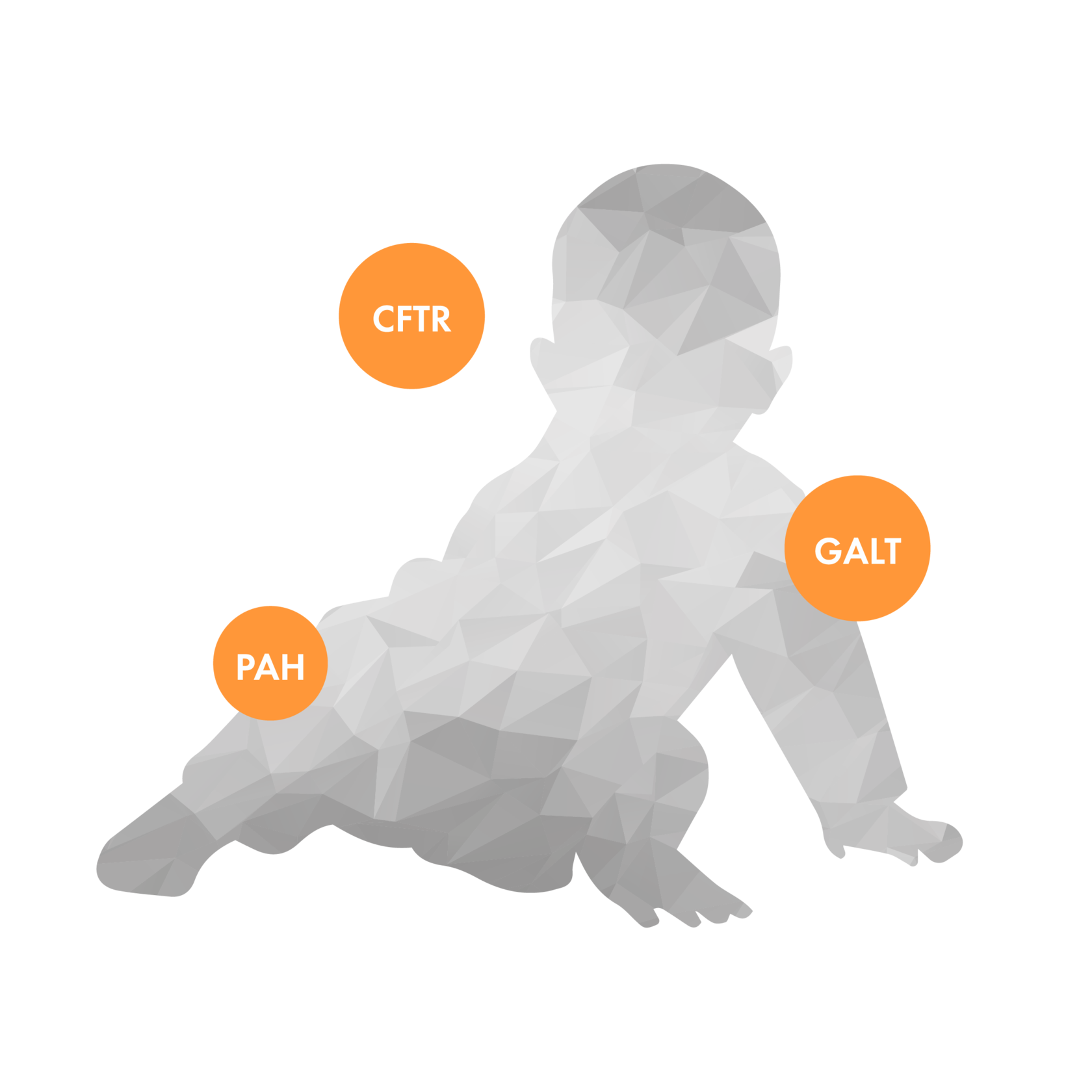
Cystic fibrosis, phenylketonuria and galactosemia are severe and relatively frequent hereditary diseases that lead to systemic disorders in the body. These monogenic diseases can be caused by hundreds of different mutations in a single gene, which makes the high-throughput sequencing method the most informative for their study. Detection of mutant variants of the CFTR, PAH and GALT genes in a child or carrier in parents allows genetic counseling for further diagnosis or reproductive manipulation.
Gene
Associated diseases
CFTR
Mucoviscidosis
GALT
Galactosemia
PAH
Phenylketonuria
Features
Specifications
Number of genes
3
Analyzed regions
Сoding DNA sequences (CDSs), exon-intron boundaries, 3'- and 5'-untranslated regions (UTRs), and some deep intronic regions that contain clinically relevant variants. The target regions of the GALT gene additionally include all intronic regions.
Limitations
The kit is not designed to detect CNV (except CFTRdele2-3), extended STR and homopolymer variants, 6 rare pathogenic variants located in deep intronic regions of the CFTR gene.
Analyte
DNA isolated from peripheral blood, dry blood spots
Number of amplicons
128 (2 pool)
Recommended number of reads per sample
64 000
Biological identifier (BID)*
Included
Time to results
from 26 to 34 hours
Compatible platforms
Illumina
Ordering information

GeneValue™ Neoscreen Solution, 24 reactions
Cat.No. GV-NEOS-24
Contents: reagent kit for targeted amplification and library preparation, access to GeneValue™ Germline Software.
Contents: reagent kit for targeted amplification and library preparation, access to GeneValue™ Germline Software.

GeneValue™ Neoscreen Solution, 48 reactions
Cat.No. GV-NEOS-48
Contents: reagent kit for targeted amplification and library preparation, access to GeneValue™ Germline Software.
Contents: reagent kit for targeted amplification and library preparation, access to GeneValue™ Germline Software.