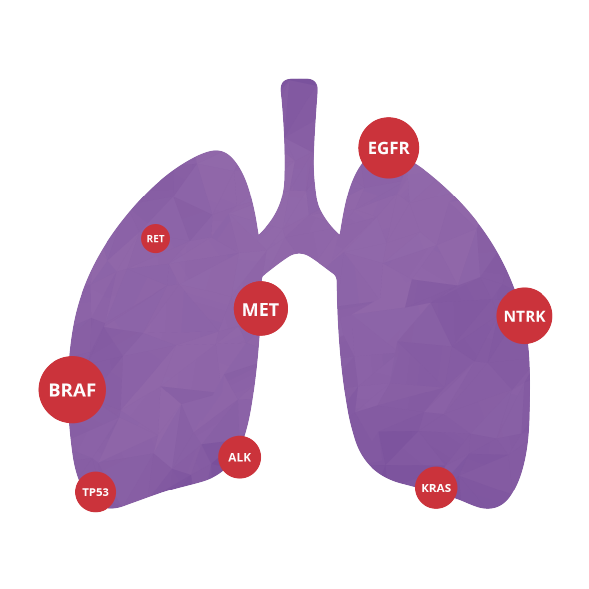
Test system for the determination of drug susceptibility or resistance markers in non-small cell lung cancer (NSCLC).
GeneValue™ NSCLC Solution
For research use only

Contains library preparation reagents, pipeline and access to GeneValue™ Somatic Software for data analysis.
Key Benefits
Two stage library preparation protocol
Simultaneous DNA and RNA analysis
Validated pipeline and the software for data analysis
AMP/ASCO/CAP and ACMG variants annotation
Time to result - 24 hours for 16 samples
GeneValue™ NSCLC Solution is designed to determinate significant genetic variants in non-small cell lung cancer (NSCLC) samples by NGS from human genomic DNA and total RNA isolated from Formalin-fixed, paraffin-embedded (FFPE).
Features
Specifications
Analyzed
DNA regions
DNA regions
All coding regions (CDSs) of EGFR, KRAS, TP53, NRAS genes;
20-25 exons ALK;
11, 15 exons BRAF;
16-21 exons ERBB2 (HER2);
2, 8, 10, 14, 21 exons PIK3CA;
hotspots of MET and ROS1genes
20-25 exons ALK;
11, 15 exons BRAF;
16-21 exons ERBB2 (HER2);
2, 8, 10, 14, 21 exons PIK3CA;
hotspots of MET and ROS1genes
Analyzed fusions
Any fusions for oncogenes: NTRK1, NTRK2, NTRK3, ALK, ROS1, RET
Gene partner indication only for partners:AKAP13, IRF2BP2, SQSTM1, TRIM24, ETV6, TPM3, EML4, CD74, KIF5B
Gene partner indication only for partners:AKAP13, IRF2BP2, SQSTM1, TRIM24, ETV6, TPM3, EML4, CD74, KIF5B
Exon skipping detection
MET exon 14 skipping;
ERBB2 exon 16 skipping
ERBB2 exon 16 skipping
Features
Specifications
Number of genes
23
Detected variants
DNA: SNV, INDEL
RNA: exon skipping, fusions
RNA: exon skipping, fusions
Limitations
The kit is not intended for detection of CNV, extended STR and homopolymer variants
Analyte
DNA and RNA isolated from formalin-fixed, paraffin-embedded tissues (FFPE)
Target enrichment
Multiplex PCR
DNA sequencing regions length, b.p.
9 135
Recommended number of reads per sample
500 000
Read Length
PE 150 + 150
Biological identifier (BID)*
Included
Time to results
24 hours
Data analysis software
Web-based GeneValue™ Software
DualStream Pipeline
GeneValue™ annotation database
DualStream Pipeline
GeneValue™ annotation database
Compatible platforms
MiSeq™, MiSeq Dx™, NextSeq™

GeneValue™ NSCLC Solution set 1 (RUO), 96 reactions
Cat.No. GV-L1-96
Contents: Reagent kit for DNA and RNA library preparation with indexes A1…H12, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A1…H12, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution set 1-1 (RUO), 16 reactions
Cat.No. GV-L11-16
Contents: Reagent kit for DNA and RNA library preparation with indexes A1…H2, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A1…H2, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution set 1-2 (RUO), 16 reactions
Cat.No. GV-L12-16
Contents: Reagent kit for DNA and RNA library preparation with indexes A3…H4, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A3…H4, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution set 1-3 (RUO), 16 reactions
Cat.No. GV-L13-16
Contents: Reagent kit for DNA and RNA library preparation with indexes A5…H6, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A5…H6, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution set 1-4 (RUO), 16 reactions
Cat.No. GV-L14-16
Contents: Reagent kit for DNA and RNA library preparation with indexes A7…H8, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A7…H8, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution set 1-5 (RUO), 16 reactions
Cat.No. GV-L15-16
Contents: Reagent kit for DNA and RNA library preparation with indexes A9…H10, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A9…H10, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution set 1-6 (RUO), 16 reactions
Cat.No. GV-L16-16
Contents: Reagent kit for DNA and RNA library preparation with indexes A11…H12, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA and RNA library preparation with indexes A11…H12, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1 (RUO), 96 reactions
Cat.No. GV-LD1-96
Contents: Reagent kit for DNA library preparation with indexes A1…H12, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A1…H12, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1-1 (RUO), 16 reactions
Cat.No. GV-LD11-16
Contents: Reagent kit for DNA library preparation with indexes A1…H2, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A1…H2, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1-2 (RUO), 16 reactions
Cat.No. GV-LD12-16
Contents: Reagent kit for DNA library preparation with indexes A3…H4, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A3…H4, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1-3 (RUO), 16 reactions
Cat.No. GV-LD13-16
Contents: Reagent kit for DNA library preparation with indexes A5…H6, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A5…H6, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1-4 (RUO), 16 reactions
Cat.No. GV-LD14-16
Contents: Reagent kit for DNA library preparation with indexes A7…H8, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A7…H8, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1-5 (RUO), 16 reactions
Cat.No. GV-LD15-16
Contents: Reagent kit for DNA library preparation with indexes A9…H10, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A9…H10, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution DNA set 1-6 (RUO), 16 reactions
Cat.No. GV-LD16-16
Contents: Reagent kit for DNA library preparation with indexes A11…H12, access to GeneValue™ Somatic Software.
Contents: Reagent kit for DNA library preparation with indexes A11…H12, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1 (RUO), 96 reactions
Cat.No. GV-LR1-96
Contents: Reagent kit for RNA library preparation with indexes A1…H12, access to
GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A1…H12, access to
GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1-1 (RUO), 16 reactions
Cat.No. GV-LR11-16
Contents: Reagent kit for RNA library preparation with indexes A1…H2, access to GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A1…H2, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1-2 (RUO), 16 reactions
Cat.No. GV-LR12-16
Contents: Reagent kit for RNA library preparation with indexes A3…H4, access to
GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A3…H4, access to
GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1-3 (RUO), 16 reactions
Cat.No. GV-LR13-16
Contents: Reagent kit for RNA library preparation with indexes A5…H6, access to
GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A5…H6, access to
GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1-4 (RUO), 16 reactions
Cat.No. GV-LR14-16
Contents: Reagent kit for RNA library preparation with indexes A7…H8, access to GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A7…H8, access to GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1-5 (RUO), 16 reactions
Cat.No. GV-LR15-16
Contents: Reagent kit for RNA library preparation with indexes A9…H10, access to
GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A9…H10, access to
GeneValue™ Somatic Software.

GeneValue™ NSCLC Solution RNA set 1-6 (RUO), 16 reactions
Cat.No. GV-LR16-16RUO
Contents: Reagent kit for RNA library preparation with indexes A11…H12, access to
GeneValue™ Somatic Software.
Contents: Reagent kit for RNA library preparation with indexes A11…H12, access to
GeneValue™ Somatic Software.